Classifying plant disease status with CNNs
Comparing simple models vs pretrained
Machine learning through Convolution Neural Networks is something I’ve been interested in for a few years, but never quite had the right data for, so instead I decided to dip my toes in with a short little project training a couple of CNNs to classify images!
For my first project I didn’t want something too simple, but I also didn’t want to get bogged down in complexity this early on, so I used a pre-existing Kaggle dataset of plant leaf images which you can find here. The dataset contains 4502 images of healthy and diseased plant leaves across 12 species, photographed against a common dark background.
The raw images were quite large, and training a NN on them would have taken an extremely long time (rough when you want to actually use your computer!). So to address this I first reprocessed the images to a more manageable 256x256. To preserve the aspect ratio, I first rescale such that the shortest side is 256px, then crop the longer side to match. Another option would be to resize without respect for the aspect ratio, although this could introduce biases if later data is introduced with a different original aspect ratio.
I then augment my training data with some rotation and scaling, and train two CNNs:
A simple model containing a few convolution layers with increasing filter size, followed by a fully-connected layer.
A pre-trained NN for which I remove the top, freeze the lower layers, and add specialised training layers for my problem.
They both do okay, although the pre-trained model reaches a higher accuracy much faster!
The simple model
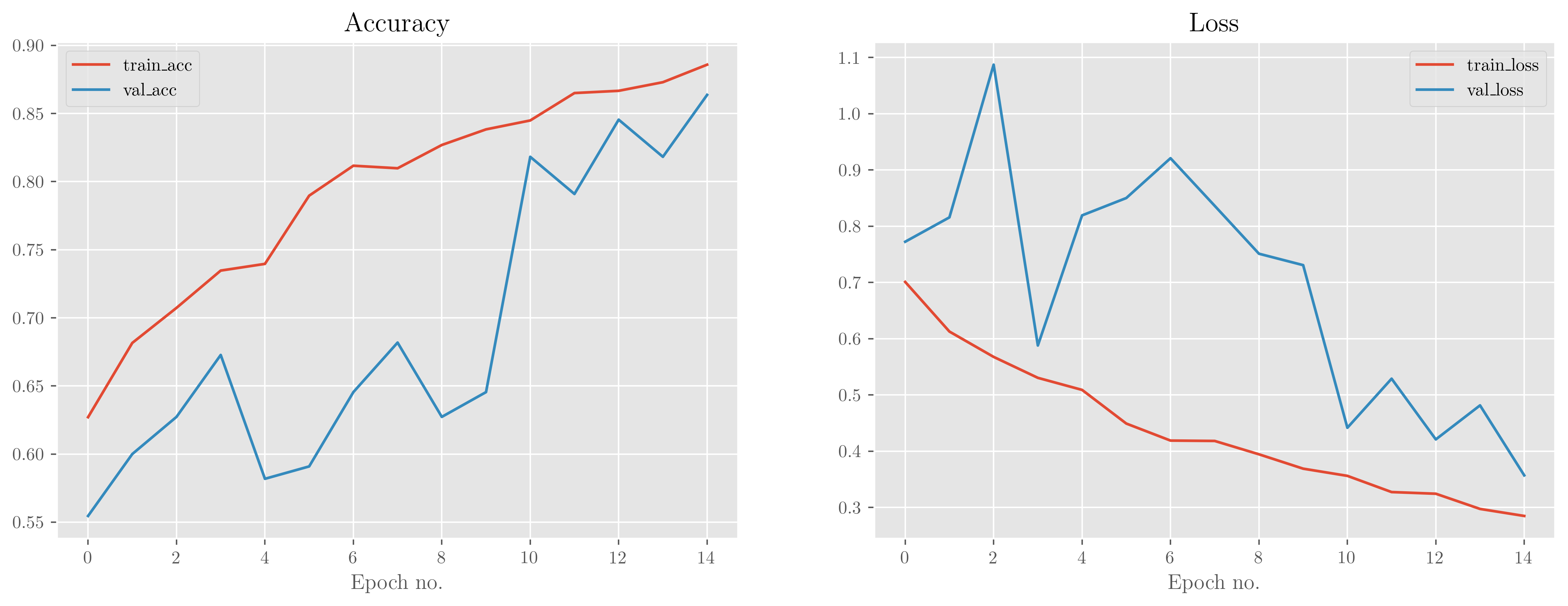
We see that the validation accuracy and loss is a little noisy but in general continues to follow the trend of the training accuracy/loss, perhaps indicating that this model could be run for even longer. Validation accuracy/loss does sit slightly below/above that of the training set, though, which suggests that the model is slightly overfitting to the training data—not unexpected given the small amount of data we have. The simple model utilises Batch Normalisation and Dropout, but these can only help so much.
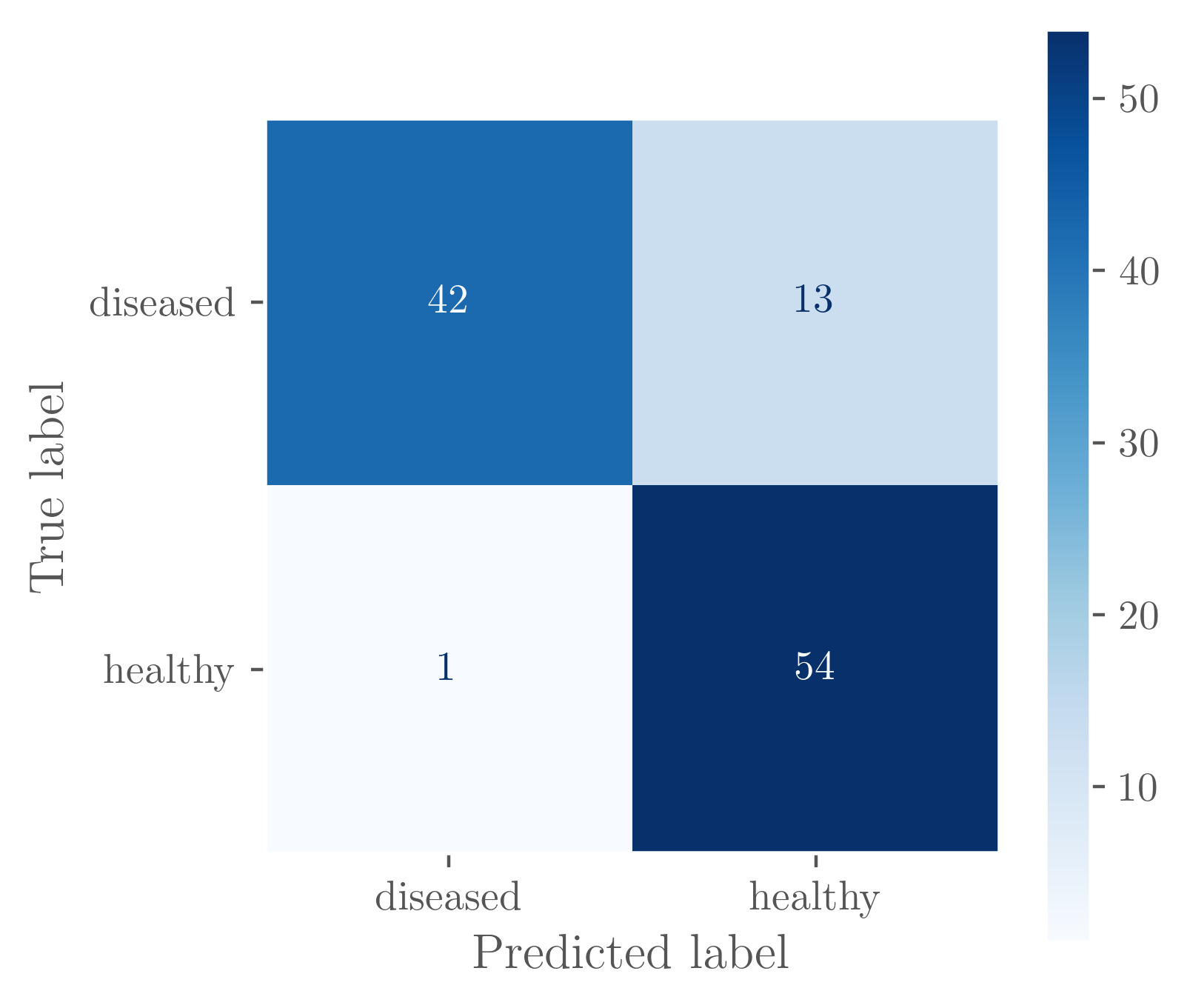
The confusion matrix also gives us some interesting insight into what the model is doing. We can see that it tends to work very well on healthy leaves, but struggles somewhat with identifying diseased ones. This might be because of the shear variation in leaf types and their corresponding diseases, i.e. a healthy leaf is smooth and green, but a diseased leaf can be brown or yellow, with bumps or smooth, intact or torn.
The pretrained model
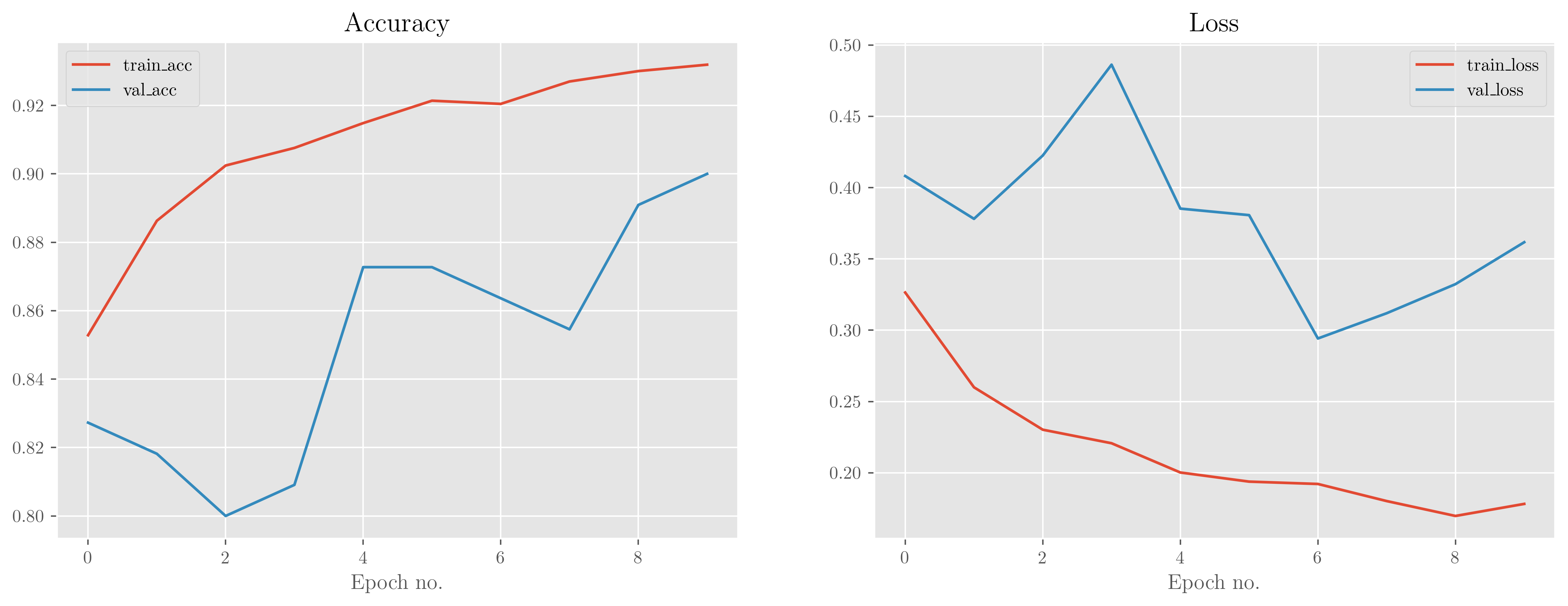
Our pretrained model does at least as well as the simple model right out of the box, and its accuracy only increases with the minimal training we provide it. Validation measures remain close to the training ones, but again may indicate a slight overfitting that is increasing in time. This is a very powerful model, though, and in general, the more complex the model the faster it will overfit.
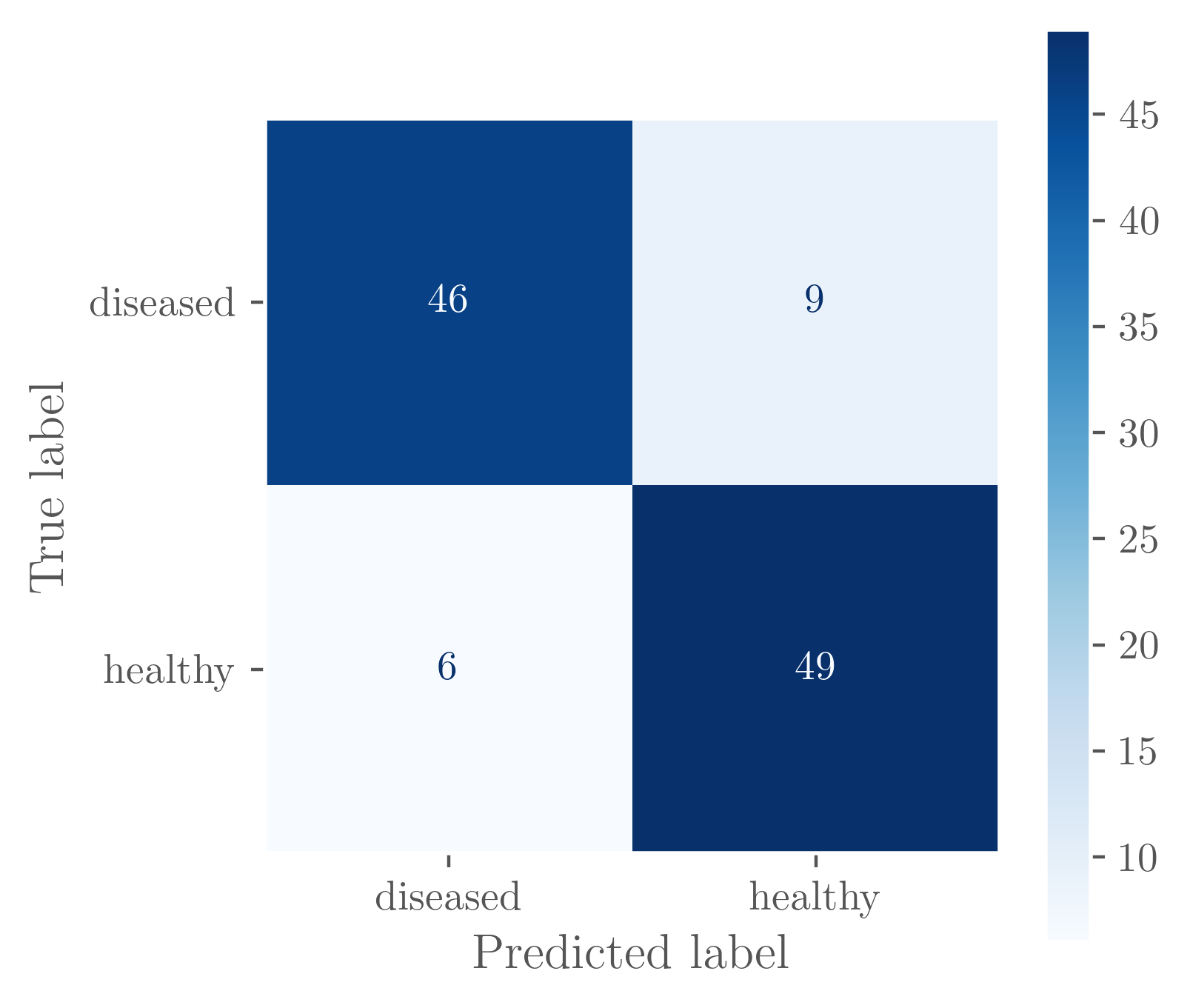
Regardless, the confusion matrix shows that this model is handling the data much better and more consistently. It doesn’t get it right all the time, but tends to identify healthy and diseased leaves with roughly the same true/false-positive/negative rates.
The takeaway
While the specific design of a model is important, at the end of the day the number and quality of the data is what really limits the accuracy of a CNN model. In some settings it will make sense to build a new CNN and train on a local dataset, but for most companies and most applications, it’s much more efficient to use a pre-existing model and train the last few layers.